Drug and Gene Screening –
Treating the UnTreatable
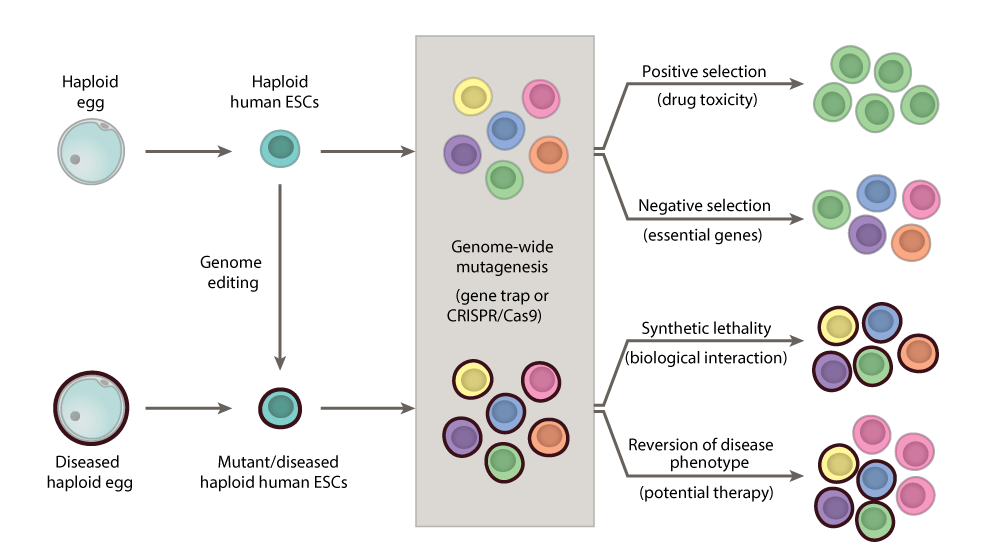
For the first time, NewStem’s Proprietary Haploid ESCs introduces expansive screenings capabilities. This advances our comprehension of human biology in health and disease as well as the identification of genes and drugs relevant for genetic and epigenetic disorders.